| Table of Contents | ||
|---|---|---|
|
WEAVE Open-Time Phase 2. Instructions for Applicants
Important links and documentation
WEAVE Automated Submission Platform (WASP): http://wasp.ast.cam.ac.uk/
...
WEAVE Open Time programme: https://www.ing.iac.es/astronomy/observing/WEAVE-OT.html
Communication with the PI
The WEAVE Automated Submission Platform (WASP) automatically emails the principal investigators (PIs) with submission deadlines, validation acknowledgements and A detailed description of how the observations are processed and eventually released can be found in the WEAVE data model documentation:
- Login to WASP or the OR.
- Click on ("Tools" and select "Data Model").
- In section "Resources", click on the "Documentation" tab and download WEAVE-ICD-030: "Interface between SPA and SWG/QAG (info for WASP etc)".
Communication with the PI
The WEAVE Automated Submission Platform (WASP) automatically emails the principal investigators (PIs) with submission deadlines, validation acknowledgements and any other important information concerning the preparation of phase 2. These The first two emails contain the words [WEAVE-WASP] at the beginning of the subject line.
Summary of actions
...
the following (the subject line is provided):
- An email containing the credentials to log onto WASP:
Subject: [WEAVE] Account created for WEAVE observation preparation - A following email contains information about the opening of the OB submission window:
Subject: [WEAVE-WASP] 20XXXX WASP upload window now open
General assistance can be provided at the following email:
weave_open_time_support@ing.iac.es |
Summary of actions
| GREEN: Actions by the PI |
| PURPLE: Actions by the PI if familiarised with the package "IFU Workflow", or by ING on behalf of the PI, if assistance is requested by the PI to weave_open_time_support@ing.iac.es |
...
The instructions below are mainly aimed at PIs who will request assistance to:
weave_open_time_support@ing.iac.es |
LIFU mode: catalogue preparation
...
WASP sets limits to allowed target RA and Dec. WASP , and it will not validate your OB is if your target coordinates lie outside the limits for that trimester. See below the allowed RAs for each trimester. The total length (counted as science exposure time) of all the OBs submitted over the two trimesters can't exceed the science time allocated to your proposal in that semester (see time allocations).
...
20h (300º) → 16h (240º)
...
declination limit is -25º.
| Trimester A1 | |||
2h (30º) → 20h (300º) Expanded range: | |||
| Trimester A2 | |||
9h (135º) → 2h (30º) Expanded range: | > -25º | ||
| Trimester A1 | |||
| OB submission | Observing period | Allowed RAs | Dec limit |
| ~weeks before observing period starts | 1 Feb - 30 Apr | 2h (30º) → 20h (300º | |
| Trimester B1 | |||
|---|---|---|---|
15h (225º) → 9h (135º) Expanded RA range: | > -25º | ||
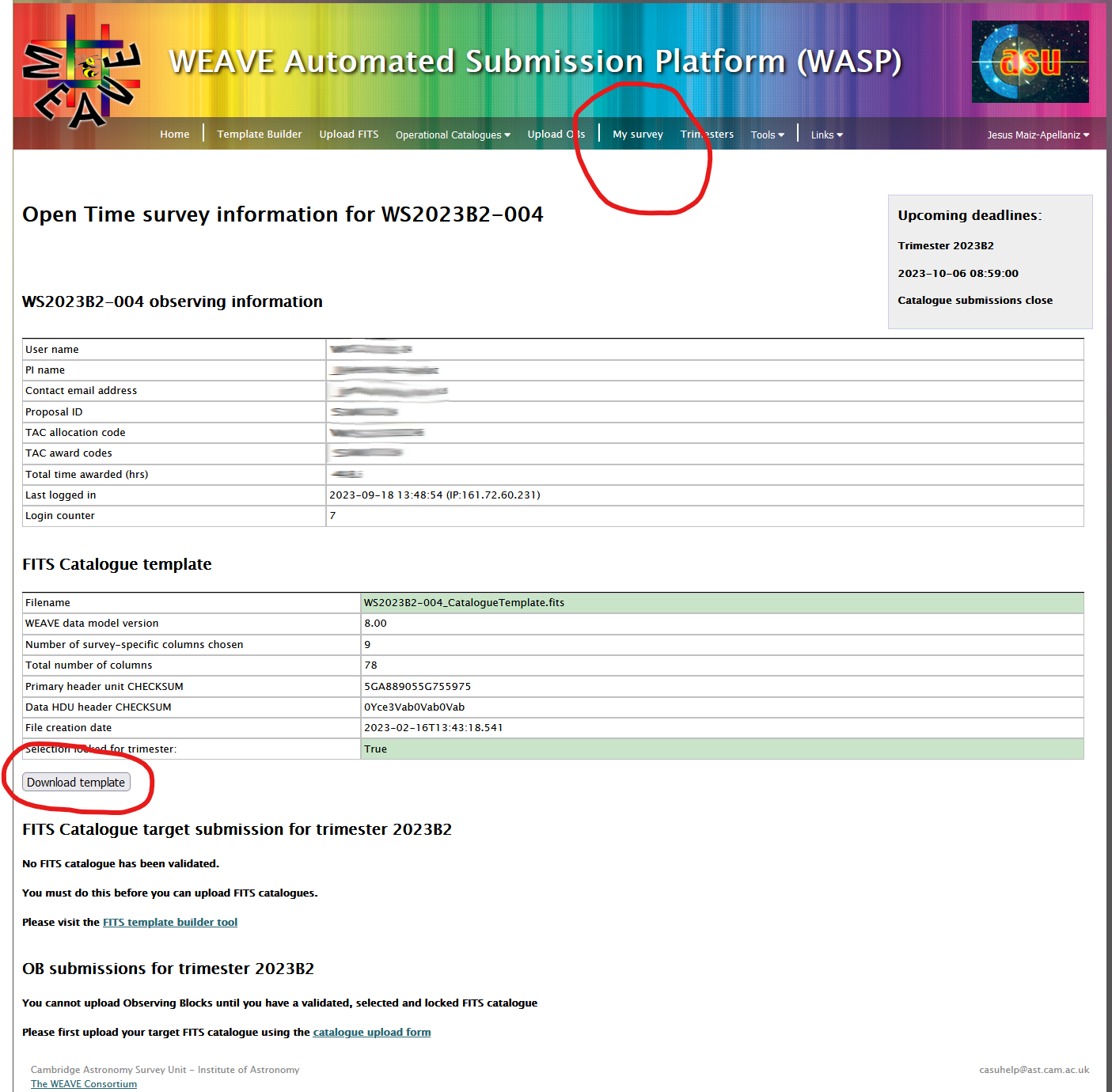
Download the catalogue template
Log on to WASP with the username and password provided in an email you must have received from WASP. Go to "My survey" and click on "Download template".
Alternative catalogue template
Alternatively, a simple template in CSV format can be used. Below it's a template with one target:
120º-135º |
| Trimester B2 |
|---|
20h (300º) → 16h (240º) Expanded RA range: |
Total requested time
The total length (counted as science exposure time) of all the submitted OBs can't exceed the science time allocation of your proposal (see the time allocations).
Preparation of the catalogue
The catalogue can be prepared in three alternative ways:
- Filling in the FITS catalogue template provided on WASP. Log on to WASP with your credentials. Go to "My survey" and click on "Download template".
2. Converting the above FITS catalogue template into CSV and filling in.
3. Filling in a simplified CSV version of the catalogue template. Below it's an example of this simplified template and one target:
<filename: WS2023B2-001_target_list.csv>TARGID,TARGNAME,GAIA_RA,GAIA_DEC,PROGTEMP,OBSTEMP,IFU_DITHER |
Filling out the catalogue template with targets
These are the columns in the catalogue template we request PIs to fill in:
...
GHMon,GHMon_NW,32.591451,23.910124,60881,IADEE,4 |
Filling out the catalogue template with targets
Any of the three catalogue templates above (FITS, CSV, simplified CSV) can be filled in with targets. These are the requested columns (the rest are optional):
TARGPROG (optional)
TARGID
TARGNAME
GAIA_ID (if available)
GAIA_RA
GAIA_DEC
GAIA_PMRA (if available)
GAIA_PMDEC (if available)
GAIA_PARAL (if available)
PROGTEMP
OBSTEMP
IFU_DITHER
IFU_PA_REQUEST (if necessary)
TARGPROG
TARGPROG is an optional column, to be filled out at the discretion of the catalogue creator.
TARGID and TARGNAME
This parameter is used to group IFU observations of the same target, in cases where stacks are required. This helps the Core Processing System (CPS) responsible for obtaining the data, to identify cases where the same astrophysical target is observed but the OBs executed were not related.
An example would be LIFU observations of the core of M33. If a user requires 3 OBs, each with different dither positions, then the CPS could not ordinarily stack these data, because they do not share the same Central CNAME (CNAME is the WEAVE object name from coordinates).
It is not always true that IFU observations with common TARGNAME will be stacked. Only common TARGNAMEs with sufficient overlap will be stacked by the CPS. However, if Contributed Data Products exist to create larger mosaics from these data, then they should use the common TARGNAME to group L1 products_PA_REQUEST (if necessary)
TARGPROG
TARGPROG is an optional column, to be filled out at the discretion of the catalogue creator.
TARGID and TARGNAME
This parameter is used to group IFU observations of the same target, in cases where stacks are required. This helps the Core Processing System (CPS) responsible for obtaining the data, to identify cases where the same astrophysical target is observed but the OBs executed were not related (via for example the "chained" directive).
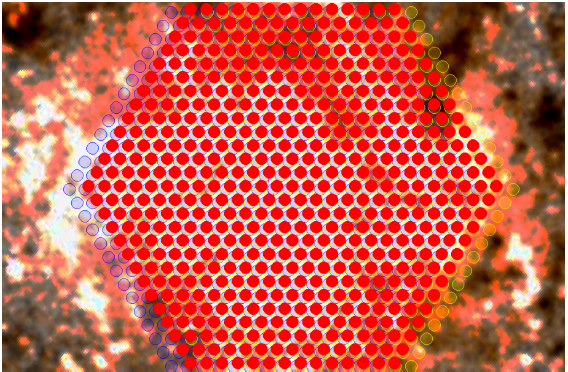
An example would be LIFU observations of the core of M33. If a user requires 3 OBs, each with different dither positions, then the CPS could not ordinarily stack these data, because they do not share the same Central CNAME (CNAME is the WEAVE object name from coordinates). In the input FITS catalogue, these observations are represented by 5,427 rows: 603 fibres, dithered 3 times for 3 OBs. Each of these rows must be tied together by a common TARGNAME, e.g. "M33 bulge". This indicates to the CPS that these observations should be evaluated for stacking.
Above: Three LIFU dither positions centred on a putative target. This information must be encoded in the input FITS catalogue, but grouped by common TARGNAME.
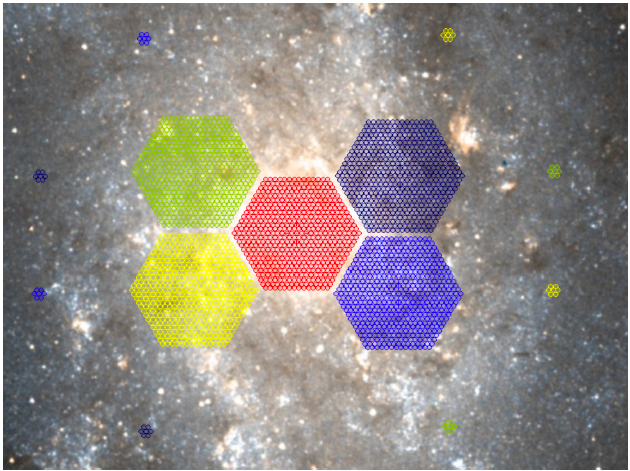
It is not always true that IFU observations with common TARGNAME will be stacked. Only common TARGNAMEs with sufficient overlap will be stacked by the CPS. However, if Contributed Data Products exist to create larger mosaics from these data, then they should use the common TARGNAME to group L1 products.
Above: Example LIFU coverage of an extended source. Each colour here represents an OB. Within each pointing there would be a series of dithers (as per previous Figure). Whilst these pointings might share the same TARGNAME ("M33"), the CPS would recognise these as a mosaic and not stack them into a single LIFU data cube.
The TARGID is the target identifier and it is mandatory that this column is filled in.
For IFU fields, this is the OB-specific descriptor of the field. An example of TARGIDs of OBs with different pointings, In the first example above, this could be a simple numerical identifier for each OB:•
OB1: TARGNAME = “M33bulge” TARGID = “M33bulge1”
...
OB2: TARGNAME = “M33bulge” TARGID = “M33bulge2”
...
OB3: TARGNAME = “M33bulge” TARGID = “M33bulge3”
But for overlapping OBsFor the second example, TARGID could be more descriptive:
•
OB1: TARGNAME = “M33” TARGID = “M33 bulge”
...
OB2: TARGNAME = “M33” TARGID = “M33 disc NE”
...
OB3: TARGNAME = “M33” TARGID = “M33 disc NW”
PROGTEMP
The PROGTEMP code is an integral part of describing how the instrument is configured. This parameter encodes the requested instrument configuration, OB length, exposure time, spectral binning, cloning requirements and probabilistic connection between these clones.
...
For custom dither patterns, constraints on the dither step size are imposed by the WASP to ensure that the guide star remains within the guiding camera field of view. From the perspective of IFU users, careful consideration of the dithering options should be made. Below it's an example of custom dithering. Note that pointings are provided for each dither, rather than the dither pointings from an initial positionthe dither pointings from an initial position. The three pointings provided in the example below would be executed as part of one only OB.
# A custom 3-dither pattern LIFU pointing |
...
| Column | Description | Format | Length | Value(s) | Units | Example | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TARGPRO | Optional description of programme | ASCII | ≤40 | WS2023B2-010_001 | |||||||||||
| TARGNAME | The target name | ASCII | ≤30 | M33 | |||||||||||
| TARGID | The identifier of the target assigned for this programme | ASCII | ≤30 | M33_NE | |||||||||||
| PROGTEMP | Observing Programme Template | ASCII | 8 (fixed) | 41331.3+ | |||||||||||
| OBSTEMP | Observing Constraints Template | ASCII | 5 (fixed) | FAACA | |||||||||||
| GAIA_DR | GAIA Data Release version | I1 | 2, 3 | 3 | |||||||||||
| GAIA_RA | Gaia RA of target | F11.7 | 0...360 | degrees | 178.221875 | ||||||||||
| GAIA_DEC | Gaia Dec of target | F11.7 | -90...90 | degrees | 44.123919 | ||||||||||
| GAIA_EPOCH | Gaia Epoch of target | F6.1 | 2015.5, 2016.0 | Julian year | 2016.0 | ||||||||||
| GAIA_PMRA | Gaia Proper Motion of target in RA | F11.3 | mas/yr | 12.1 | |||||||||||
| GAIA_PMDEC | Gaia Proper Motion of target in Dec | F11.3 | mas/yr | 0.01 | |||||||||||
| GAIA_PARAL | Gaia Parallax of target | F10.3 | mas | 0.002 | IFU_PA_REQUEST | Position Angle of IFU bundle | F11.7 | -180...180 | degrees | 106.701 | IFU_DITHER | IFU dither pattern code | I2 | -3, -1, 0, 3, 4, 5, 6 | 3 |
Below it's an example of a CSV catalogue file:
| _PARAL | Gaia Parallax of target | F10.3 | mas | 0.002 | ||
| IFU_PA_REQUEST | Position Angle of IFU bundle | F11.7 | -180...180 | degrees | 106.701 | |
| IFU_DITHER | IFU dither pattern code | I2 | -3, -1, 0, 3, 4, 5, 6 | 3 |
Aladin overlays of the LIFU footprints
...
The edited catalogue must go through the IFU workflow, a set of scripts which will make a version of the FITS catalogue suitable for uploading to WASP (http://wasp.ast.cam.ac.uk/catsubmit/upload) for validation. First log on using the provided credentials (the ones emailed from WASP), note that you need to register your IP at http://wasp.ast.cam.ac.uk/ipsubmit/upload first. See below the tab used for uploading the catalogue.
...
At ING, we can provide assistance in guiding you through the different stages of the IFU workflow package, or we can even process your catalogue provided in any of the formats described above, on your behalf. Please contactIn this latter case, simply email your catalogue to the contact below:
weave_open_time_support@ing.iac.es |
...
At ING, we can provide assistance in generating the OBs from the CNAMEd catalogue, please contact:
weave_open_time_support@ing.iac.es |
OB upload to WASP
The processed OBs can be then uploaded, as a tar file if many, to http://wasp.ast.cam.ac.uk/xmlsubmit/upload (see below). Remember to log on first.
...